Research Background: To date, about 500 primary immunodeficiencies are known, but the list is growing yearly as new diseases emerge. Although individually rare, the overall incidence of primary immunodeficiency is estimated at 1 in 10,000 individuals [1]. A collaborative research team from the LKS Faculty of Medicine of the University of Hong Kong and the Centre for Translational Stem Cell Biology has pioneered a new stem cell model to help personalise treatment for patients suffering from rare forms of immunodeficiency. The research findings were published in the Journal of Allergy and Clinical Immunology.
- Front Immunol. 2021 Feb 18:11:625753. doi: 10.3389/fimmu.2020.625753.
Translational Stem Cell Research Bringing Hope to Patients with Rare Diseases
Primary immunodeficiencies, also referred to as 'inborn errors of immunity' (IEI), encompass a wide array of disorders that impair the immune system. These are rare disorders that often face under-diagnosis and lack of awareness, leading to significant challenges for affected individuals. This impairment leaves individuals extremely vulnerable to infections, autoimmunity, and even cancer [1]. Despite their rarity on an individual level, IEI contribute significantly to illness and death when considered collectively. The importance of relatively under-studied research in this field cannot be overstated, as it aims to enhance our understanding of these complex disorders, improve diagnostic techniques, and develop tailored therapeutic approaches.
One such IEI, known as Signal transducer and activator of transcription (STAT)-1 gain of function (GoF), was first documented in 2011 as autosomal-dominant chronic mucocutaneous candidiasis (CMC) [2]. This disorder presents a diverse clinical phenotype, with patients in the same family, carrying the same mutation, experiencing varying disease manifestations and severity [3,4,5]. This variance is likely due to the differing underlying mechanisms of STAT1-GoF in different mutations. However, studying and confirming the function of individual mutations has proved challenging due to the limited availability of fresh patient samples. Thus, there is a need for a specific disease model that can mimic the patient-specific and mutation-specific properties of STAT1-GoF, eliminating the need for repeated sampling.
Traditionally, many IEI, including STAT1-GoF, are managed with antimicrobial suppression and immunosuppression for autoimmune manifestations. However, these treatments are often accompanied by numerous side effects and poor outcomes. Hematopoietic stem cell transplantation for STAT1-GoF, despite its potential for a cure, has high failure rates [6]. Moreover, while the recent use of Janus activating kinase (JAK) inhibitors (JAKi) has shown promising effects in treating STAT1-GoF, the complexity of STAT1-GoF pathophysiology and patient heterogeneity means that patients are likely to respond differently to discrete JAKi depending on the underlying disease mechanism [7,8]. This underscores the need for ex-vivo and patient-specific testing platforms for the evaluation of novel therapies.
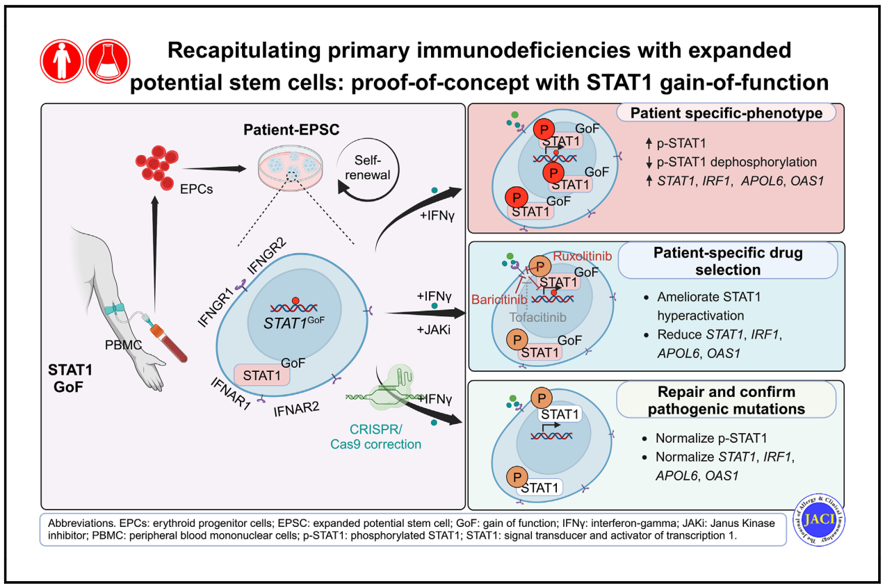
In our study, we explored the potential of patient-derived expanded potential stem cells (EPSC) as an ex-vivo platform for disease modelling and personalised treatment. We generated EPSC cell lines derived from individual STAT1-GoF patients and healthy controls. EPSC, a type of expanded potent stem cell that can be reprogrammed from somatic cells, possess superior developmental potency for all cell lineages [10] and exhibit high proliferation rates and better genetic and epigenetic stability. This makes EPSC an ideal platform for human disease modelling and gene editing [11].
We successfully reprogrammed STAT1-GoF EPSC from erythroid progenitor cells, demonstrating comparable levels of critical pluripotent marker genes as controls. Functional tests revealed an increase in phosphorylated STAT1 (p-STAT1) and increased expression of STAT1 and its downstream genes in STAT1-GoF EPSC upon IFN-γ stimulation. This suggests that patient-derived EPSC recapitulated the functional abnormalities of index STAT1-GoF patients.
We further investigated differential effects of specific JAKi during EPSC stimulation. We found differential patient-specific responses to different JAKi among different STAT1-GoF EPSC. In particular, ruxolitinib and baricitinib inhibited STAT1 hyperactivation in STAT1-GoF EPSC in a dose-dependent manner, which was not observed with tofacinib. This result mirrors the real-life experience of one of our patients, whose disease responded to baricitinib but failed to respond to more JAK1-selective agents. This platform thus paves the way for the development of personalised treatment.
Additionally, we demonstrated functional correction of STAT1 phosphorylation and downstream gene expression in repaired STAT1-GoF EPSC. Pathogenic STAT1-GoF mutation in patient-derived EPSC lines were successfully repaired using CRISPR/Cas9 editing. Functional tests were repeated on these repaired EPSC lines, with results showing significantly lower expression of p-STAT1, STAT1, and its downstream genes after IFN-γ stimulation. By tailoring patient-specific repairs, this has brought us one step closer towards tailored gene therapy as a cure for STAT1-GoF.
This study serves as the first proof-of-concept demonstrating the feasibility of our patient-derived EPSC platform for modelling IEI. We anticipate other novel uses of this platform in future prospective trials, in addition to personalised disease modelling. For instance, by leveraging established protocols to generate organoids from stem cell lines, this would enable us to understand the differential effects of distinct STAT1 mutations on specific tissues or organs, model interactions between different cell types, and screen for therapies for different disease manifestations using patient-derived organoids. Studies inducing pathogenic mutations into healthy EPSC to compare between patient-derived and gene-edited EPSC to discover potential regulatory elements or roles of other loci are also currently underway. Ultimately, we hope to expand such platforms to other IEI and rare inherited disorders to benefit these underserved patients in the future.
References:
- P.P. Lee, H. Mao, W. Yang, K.W. Chan, M.H. Ho, T.L. Lee, et al. Penicillium marneffei infection and impaired IFN-gamma immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations J Allergy Clin Immunol, 133 (2014), pp. 894-896.e5
- P.H. Li, P.P. Lee, S.L. Fung, C.S. Lau, Y.L. Lu Chronic mucocutaneous candidiasis—more than just skin deep Hong Kong Med J, 24 (2018), pp. 423-425
- S. Giovannozzi, V. Lemmens, J. Hendrix, R. Gijsbers, R. Schrijvers Live cell imaging demonstrates multiple routes toward a STAT1 gain-of-function phenotype Front Immunol, 11 (2020), p. 1114
- S. Giovannozzi, J. Demeulemeester, R. Schrijvers, R. Gijsbers Transcriptional profiling of STAT1 gain-of-function reveals common and mutation-specific fingerprints Front Immunol, 12 (2021), Article 632997
- J. Zheng, F.L. van de Veerdonk, K.L. Crossland, S.P. Smeekens, C.M. Chan, T. Al Shehri, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC) Eur J Immunol, 45 (2015), pp. 2834-2846
- J.W. Leiding, S. Okada, D. Hagin, M. Abinun, A. Shcherbina, D.N. Balashov, et al. Hematopoietic stem cell transplantation in patients with gain-of-function signal transducer and activator of transcription 1 mutations J Allergy Clin Immunol, 141 (2018), pp. 704-717.e5
- R. Mossner, N. Diering, O. Bader, S. Forkel, T. Overbeck, U. Gross, et al. Ruxolitinib induces interleukin 17 and ameliorates chronic mucocutaneous candidiasis caused by STAT1 gain-of-function mutation Clin Infect Dis, 62 (2016), pp. 951-953
- O. Zimmerman, B. Rosler, C.S. Zerbe, L.B. Rosen, A.P. Hsu, G. Uzel, et al. Risks of ruxolitinib in STAT1 gain-of-function-associated severe fungal disease Open Forum Infect Dis, 4 (2017), p. ofx202
- X Liu, VSF Chan, KGC Smith, C Ming, CS Or, FTW Tsui, et al. Recapitulating primary immunodeficiencies with expanded potential stem cells: Proof of concept with STAT1 gain of function J Allergy Clin Immunol, 4 (2024), pp. 1125-1139.
- X. Gao, M. Nowak-Imialek, X. Chen, D. Chen, D. Herrmann, D. Ruan, et al. Establishment of porcine and human expanded potential stem cells Nat Cell Biol, 21 (2019), pp. 687-699
- Y. Yang, B. Liu, J. Xu, J. Wang, J. Wu, C. Shi, et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency Cell, 169 (2017), pp. 243-257.e25
Authors:
Prof Philip H Li, Principal Investigator, Centre for Translational Stem Cell Biology
Dr Xueyan Liu, Post-doctoral Fellow, Centre for Translational Stem Cell Biology
Prof Chak-Sing Lau, Programme Leader, Centre for Translational Stem Cell Biology
Prof Pengtao Liu, Managing Director, Centre for Translational Stem Cell Biology
September 2024