香港理工大學對2019年冠狀病毒的流行病學研究
冠狀病毒2019-20(COVID-19,新冠肺炎)大流行已經對全球公共衛生,世界經濟和社會經濟格局造成了巨大影響。香港理工大學一個研究小組投身於新冠肺炎流行病學研究的前沿。該小組的成員包括香港理工大學應用數學系的何岱海博士和樓一均博士及護理學院的楊琳博士。該小組不懈地致力於新冠肺炎的資料分析和模型分析,旨在向決策者和公眾提供最新的準確資訊。
該團隊是最活躍的流行病學研究團隊之一,並取得了一些有見地的結果。比如,該小組的關於爆發早期階段中國湖北武漢市新冠肺炎基本再生數的初步估計1的文章已在Google學術搜索中被引用418次,這表明該研究的重要性。
這是其工作的簡短摘錄。有關各種研究發現的詳細資訊,請參閱本文末尾的參考文獻。
香港理工大學的研究發現
準確估計新冠肺炎的主要流行病學參數2對於緩解爆發至關重要。香港理工大學的研究小組基於公開的資料最早(1月23日)計算出新冠肺炎的傳播性(基本再生數),即平均一個原發病例可導致2-3個繼發病例。值得一提的是,團隊首次指出確診率(報告率,報告病例佔實際感染的比例)對基本再生數的估計的影響1。每日新增的報告病例在1月中下旬突然增加,部分原因是測試能力的提高。在後續的工作中,團隊計算了1月中旬武漢市由於測試能力不夠和意識不足而漏報的病例數目3。這些工作對於認識疫情規模是非常重要的。
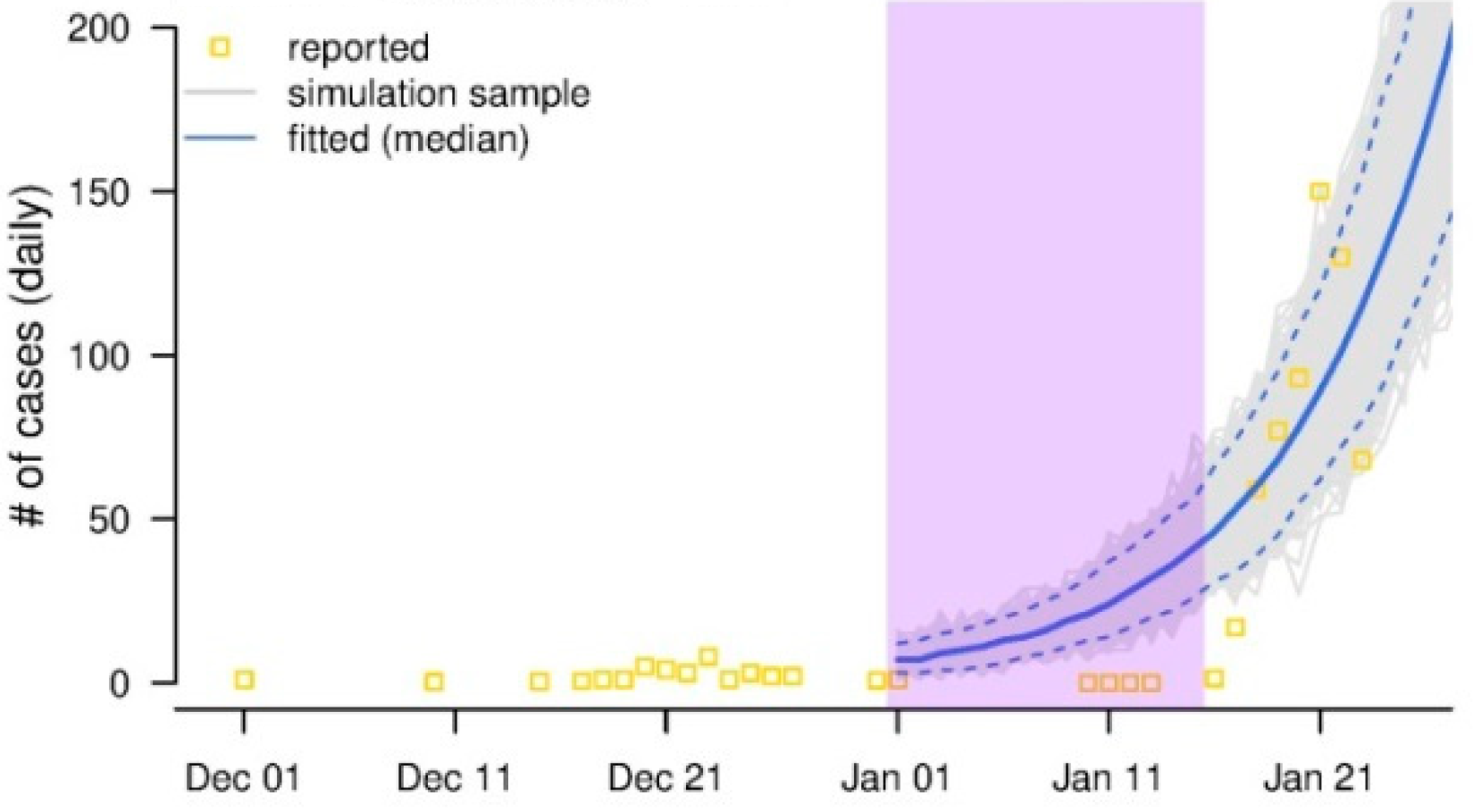
圖. 黃色方塊表示每日報告新增病例。經典的流行病學理論告訴我們,爆發中每日新增病例會呈指數增長,如藍色曲線所示。紫色陰影所示的時間間隔中,每日報告新增病例明顯低於理論預測(藍色曲線),極有可能存在漏報。通過比較藍色曲線(理論預測)和實際報告的病例,團隊估計了這段時間內的漏報3。
同時,團隊量化了交通運輸4, 5(尤其是火車運輸)對新冠肺炎從武漢市到其他大陸城市的空間擴散的影響。團隊使用簡單的線性回歸模型,基於武漢市以外城市的報告病例,以及從武漢市到其他城市的人口流動資料,估計了交通運輸對武漢市到其他城市的病例輸出的影響。團隊還量化了無症狀病例的相對傳播能力,發現無症狀病例的傳播能力是有症狀病例的1/46。因此,無症狀病例在新冠肺炎傳播中的作用可能較弱。
團隊還計算了代間隔7, 8(從原發病例症狀發作至繼發病例症狀發作的時間延遲)這一個重要參數,與基本再生數一起控制新冠肺炎的傳播速度。基本再生數決定了每個原發病例的預期繼發病例的數目,而較短的代間隔意味著一旦有了原發病例,繼發病例會在短時間內出現。因此及時發現和隔離病患和密切接觸者至關重要。
團隊是第一個量化新冠肺炎病例的傳播能力的不均勻性9,發現新冠肺炎的傳播能力不均勻性明顯小於嚴重急性呼吸系統綜合症(SARS),這部分解釋了為什麼新冠肺炎可能會長期存在並且難以根除。病例的傳播能力較均勻的病毒較容易在人群中存在。在2003年香港的SARS爆發期間,患者的傳染能力表現出較大的差異,即少數病例具有很高的傳染性(所謂的超級傳播者),從而導致許多繼發性病例,而大多數病例傳染性低,只導致了很少的繼發性病例。這使得SARS相對易於控制,即以超級傳播者為目標的控制措施有效。在新冠肺炎疫情中,雖然發生了超級傳播事件,但病例的傳染性相對均勻,因此,在沒有嚴格控制措施的情況下,新冠肺炎較容易在人群中持續傳播。
團隊開發了數學模型來評估控制措施的影響並預測中國武漢市的暴發趨勢10, 11。團隊對比1918-19年流感大流行與新冠肺炎疫情12。鑒於1918-19年流感和新冠肺炎在傳播率和嚴重程度上較相似,我們認為前者的多波特徵和持續時間可作為後者的參考。
團隊與深圳市疾病預防控制中心合作,量化 COVID-19的復發率(出院/康復的新冠肺炎患者中在隨訪中再出現RT-PCR檢測陽性的比例)13。在兩次相鄰的RT-PCR測試均為陰性並在無症狀的情況下,新冠肺炎患者可以出院。但是,團隊發現10.5%出院的患者在出院後平均4.7天左右再出現RT-PCR陽性,顯示有排出病毒(或病毒基因片斷)。這有可能是由於測試準確性,或者反映患者體內病毒的代謝過程。團隊首次量化了復發率和相關風險因素。
楊琳博士及合作者還發表了有關新冠肺炎對孕婦和新生兒的影響14,以及新冠肺炎對於醫護人員的心理健康,危險因素和社交媒體使用方面的工作15。這些都是非常重要的課題。
團隊的工作得到香港研究資助局和阿里巴巴(中國)有限公司合作研究基金的支持。
參考資料
何岱海博士、樓一均博士和楊琳榑士發佈的新冠肺炎出版刊物和預印本
- Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, Lou Y, Gao D, Yang L, He D, and Wang MH. (2020) Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. International Journal of Infectious Diseases. 01-050.
- Li Ying-ke, Zhao Shi, Lou Yi-jun, Gao Dao-zhou, Yang Lin, He Dai-hai (2020) Epidemiological parameters and models of coronavirus disease 2019 (COVID-19). Acta Physica Sinica, accepted.
- Zhao S, Musa SS, Lin Q, Ran J, Yang G, Wang W, Lou Y, Yang L, Gao D, He D, and Wang MH. (2020) Estimating of the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of January 2020: A data-driven modelling analysis of the early outbreak. Journal of Clinical Medicine. 9(2), 388.
- Zhao S, Zhuang Z, Cao P, Ran J, Gao D, Lou Y, Yang L, Cai Y, Wang W, He D, and Wang M. (2020) Quantifying the association between domestic travel and the exportation of novel coronavirus (2019-nCoV) cases from Wuhan, China in 2020: A correlational analysis. Journal of Travel Medicine. 2020:1-3.
- Zhao S, Zhuang Z, Ran J, Lin J, Yang G, Yang L and He D. (2020) The association between domestic train transportation and novel coronavirus outbreak in China, from 2019 to 2020: A data-driven correlational report. Travel Medicine and Infectious Diseases. 33:101586.
- He D, Zhao S, Lin Q, Zhuang Z, Cao P, Wang MH, and Yang L (2020). The relative transmissibility of asymptomatic COVID cases among close contacts. International Journal of Infectious Diseases. In press.
- Zhao S, Cao P, Gao D, Zhuang Z, Cai Y, Ran J, Chong MKC, Wang K, Lou Y, Wang W, Yang L, He D, and Wang MH (2020) Serial interval in the estimation of reproduction number of the novel coronavirus disease (COVID-19) during the early outbreak. Journal of Travel Medicine. In press.
- Zhao S, Cao P, Chong MKC, Gao D, Lou Y, Ran J, Wang K, Wang W, Yang L, He D, and Wang MH. (2020) COVID-19 and gender-specific difference: Analysis of public surveillance data in Hong Kong and Shenzhen, China, from January 10 to February 15, 2020. Infection Control & Hospital Epidemiology. In press.
- He D, Zhao S, Xu X, Zhuang Z, Cao P, Wang M H, Lou Y, Wu Y & Yang L (2020). Individual variation in infectiousness of coronavirus 2019 implies difficulty in control. Available at SSRN 3559370.
- Lin Q, Zhao S, Gao D, Lou Y, Yang S, Musa SS, Wang MH, Cai Y, Wang W, Yang L, He D. (2020) A conceptual model for the outbreak of coronavirus disease 2019 (COVID-19) in Wuhan, China with individual reaction and governmental action. International Journal of Infectious Diseases. 93:211-216.
- Zhao S, Stone L, Gao D, Musa SS, Chong MKC, He D, and Wang MH (2020) Imitation dynamics in the mitigation of the novel coronavirus disease (COVID-19) outbreak in Wuhan, China from 2019 to 2020. Annals of Translational Medicine. In press.
- [1] He D, Zhao S, Li Y, Zhuang Z, Cao P, Gao D, ... & Yang L (2020). History is the best model–a comparison of the COVID-19 with the 1918-19 pandemic influenza in United Kingdom. Available at SSRN 3582393.
- Tang X, Zhao S, He D, Yang L, Wang MH, Li Y, Mei S & Zou X (2020). Positive RT-PCR tests among discharged COVID-19 patients in Shenzhen, China. Infection Control & Hospital Epidemiology1-7.
- Li N, Han L, Peng M, Lv Y, Ouyang Y, Liu K, ... & Yang L (2020). Maternal and neonatal outcomes of pregnant women with COVID-19 pneumonia: a case-control study. Clinical Infectious Diseases.
- Ni M Y, Yang L, Leung C M, Li N, Yao X I, Wang Y, ... & Liao Q (2020). Mental health, risk factors, and social media use during the COVID-19 epidemic and cordon sanitaire among the community and health professionals in Wuhan, China. JMIR Public Health and Surveillance.
2020年5月